Decoding Protein Therapeutics: The Role of Biophysical Characterization Methodologies
February 13, 2024
Biophysical characterization of proteins is a necessary step in the production and development of protein therapeutics, as it allows for a comprehensive understanding of a protein’s function and structure. By analyzing and defining the higher-order structure of potential drug targets, this process serves to identify and mitigate issues that could significantly impact the efficacy and safety of a product, such as poor stability and aggregation.1,2
Moreover, biophysical characterization is a key tool in early drug discovery stages, facilitating the identification and evaluation of potential drug targets and their interaction with therapeutic compounds. With its critical insights into the structural dynamics of protein therapeutics, biophysical characterization plays a key role in optimizing the drug design process and ensuring that potentially life-saving treatments are developed with safety and efficacy in mind.3
Common Biophysical Characterization Techniques
When it comes to biophysical methods for protein characterization, there are many options available for researchers to analyze the secondary and tertiary structure of a molecule, as well as aggregation.
Secondary and tertiary structure
Differential scanning calorimetry (DSC)
This widely recognized analytical technique can measure the molar heat capacity of samples as a function of temperature. It has proven to be particularly useful in evaluating the thermal stability of biomolecules, including proteins. Despite its simplicity and speed in determining the thermal stability of biologics, DSC has limitations. The process of baseline subtraction can introduce human error, leading to variations in results. Additionally, differential scanning calorimeters have a minimum concentration limit that may not always be achievable at the bulk manufacturing scale. Lastly, the enthalpy change (ΔH) for irreversible thermal denaturation, which isn’t absolute, may result in misleading interpretations of the Gibbs free energy change (ΔG), a key indicator of protein stability.4
Circular Dichroism (CD)
This technique allows for quick evaluation of the secondary structure, folding characteristics, and binding properties of proteins. Essentially, circular dichroism involves the differential absorption of left-handed and right-handed circularly polarized light. Furthermore, circular dichroism can be utilized to investigate protein interactions and to evaluate protein thermal denaturation and ligand binding. However, these applications are considered low throughput methods, indicating that they may not be suitable for large-scale or high-volume studies.4,5
Fourier Transform Infrared (FTIR) Spectroscopy
This non-invasive technique for protein structure analysis uses minimal photon energies to prevent unwanted reactions. It avoids sample heating, requires small amounts of sample, and doesn’t necessitate high purity. Its intrinsic time resolution is a few picoseconds. However, it lacks bond selectivity, leading to overlapping protein contributions.6
Nuclear Magnetic Resonance (NMR) Spectroscopy
This is a non-destructive method for studying the structure of molecules at the atomic level, offering insights into molecular dynamics and interactions. However, it does have its limitations. These include low sensitivity to inadequate sample concentrations resulting in poor spectra, high costs associated with the instruments and their maintenance due to the need for powerful magnets and cooling liquids, and difficulty in analyzing higher molecular weight molecules due to complex spectrum interpretation.7
Biologics Aggregation
Size Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
As a precise and adaptable biophysical method for protein characterization and aggregates, this method is capable of determining the composition, mass, and oligomeric state of membrane proteins in detergent solutions. While widely adopted, SEC-MALS does have some limitations. For instance, it’s a low throughput technique that only separates proteins based on size. Moreover, quantitative analysis can only be conducted for well-separated peaks. This means if particles in solution, like the PDC or others, have a similar size to the detergent-free micelles, the data cannot be analyzed correctly.8
Analytical Ultracentrifugation (AUC)
This is a technique that’s highly effective in detecting aggregates and sensitive to the molecular shape of proteins. It’s often employed to determine the oligomerization state of proteins, estimate dissociation constants, and assess the impact of buffers on protein solubility. Despite its wide usage and accurate results, AUC is a time-intensive process. Therefore, it’s often used to confirm data obtained from high-throughput screening methods rather than as a primary method of analysis.8
Dynamic Light Scattering (DLS)
This method is used to analyze the size distribution profile of tiny particles in suspension or polymers in solution, including protein aggregates. It can be applied to membrane proteins and is capable of high-throughput analysis. However, it isn’t always the most effective tool for characterization. A limitation of DLS is its potential failure to detect aggregates in the subvisible range. Also, data interpretation can become challenging when samples are polydisperse or contain larger aggregates. A common practice to enhance data quality is to filter samples prior to testing, but this approach simply removes the larger particles, thereby evading their analysis altogether. 2,9
Membrane Microscopy
This technique can help shorten the time and sample volume requirements needed for adequate testing. For example, light obscuration (LO) — passing a stream of particles between a light source and detector — is a common protocol that was adapted from the aviation sector (which used it for fuel analysis) by the biopharmaceutical industry for differentiating particles in drugs. But, due to its high-volume requirement, observing particles in the early stages of development is not possible with LO. In addition, biologics are nearly invisible in liquid when imaged because the biologics and the media that they are in have the same refractive index. In addition, LO does not enable individual particle identification, leaving drug developers either scratching their heads about where the particles came from or inaccurately characterizing the drugs’ therapies. 10
A Hidden Threat to Drug Development Requiring Biophysical Characterization of Proteins and Aggregation
Protein aggregation is a critical quality attribute (CQA) that can have significant impacts on the efficacy and safety of biotherapeutics. Assessing, formulating, monitoring, and mitigating protein aggregation requires in-depth analysis of various parameters, such as size, count, morphology, shape, and identification. These biophysical methods for protein characterization can be complex, time-consuming, and resource-intensive, requiring specialized expertise and equipment.
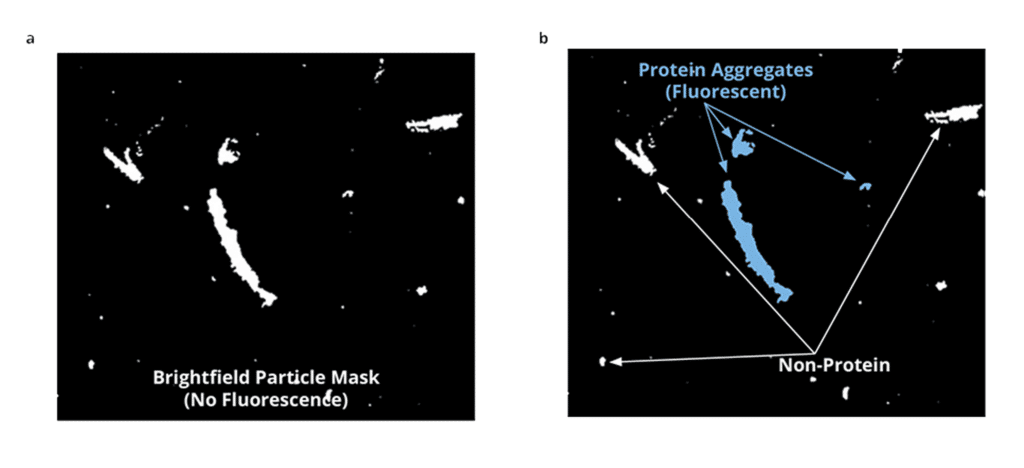
Fortunately, with Aura®’s advanced analysis capabilities, powered by Backgrounded Membrane Imaging (BMI) and Fluorescence Membrane Microscopy (FMM) technology, clear and fast particle analysis data is possible.
FMM is an innovative particle identification method that utilizes BMI to detect and categorize the most common particles in bioformulation samples. This advanced biophysical characterization technique leverages BMI, which captures images of a 96-well membrane plate both prior to and after sample filtration, using high optical contrast image analysis to distinguish particles ranging in size from 1 μm to 5 mm, with a dynamic range of over 36/mL counts. FMM then utilizes established extrinsic fluorescent dye chemistries to analyze the particles and confirm their presence. By staining particles and extracting vital information about their count, morphology, sizing, and light scattering intensity from the particle mask generated by the software, FMM offers an invaluable method for scrutinizing complex biological samples.
Conclusion
Historically, legacy biophysical methods for protein characterization tools have demanded significant time and sample volumes and could only be used in the final stages of development, which is already too late if a drug is ineffective or potentially harmful. However, innovative platforms like Aura that leverage techniques such as contemporary forms of membrane microscopy are shortening the time and sample volume requirements necessary for adequate testing.10
This technology is driving decision-making during late discovery and throughout development, parts of the process where conventional particle analyzer technology is too cumbersome and sample-consuming to use.
References
- Solomon, T.L., Delaglio, F., Giddens, J.P., Marino, J.P., Yu, Y.B., Taraban, M.B., Brinson, R.G. “Correlated analytical and functional evaluation of higher order structure perturbations from oxidation of NISTmAb.” MAbs, vol. 15, no. 1, 2023, pp. 2160227. PMC9872951. doi:10.1080/19420862.2022.2160227
- Vedadi, M., Arrowsmith, C.H., Allali-Hassani, A., Senisterra, G., Wasney, G.A. “Biophysical characterization of recombinant proteins: a key to higher structural genomics success.” J Struct Biol, vol. 172, no. 1, 2010, pp. 107-19. PMC2954336. doi:10.1016/j.jsb.2010.05.005
- Kwan, T.O.C., Reis, R., Siligardi, G., Hussain, R., Cheruvara, H., Moraes, I. “Selection of Biophysical Methods for Characterisation of Membrane Proteins.” Int J Mol Sci, vol. 20, no. 10, 2019, p. 2605. PMC6566885. doi:10.3390/ijms20102605
- Durowoju, I.B., Bhandal, K.S., Hu, J., Carpick, B., Kirkitadze, M. “Differential Scanning Calorimetry – A Method for Assessing the Thermal Stability and Conformation of Protein Antigen.” J Vis Exp, no. 121, 2017, p. 55262. PMC5409303. doi:10.3791/55262
- Vedadi, M., Arrowsmith, C.H., Allali-Hassani, A., Senisterra, G., Wasney, G.A. “Biophysical characterization of recombinant proteins: a key to higher structural genomics success.” J Struct Biol, vol. 172, no. 1, 2010, pp. 107-19. PMC2954336. doi:10.1016/j.jsb.2010.05.005
- “News Medical.” The Role of FTIR in Protein Analysis and Biomedical Applications, 18 July 2022, https://www.news-medical.net/news/20220718/The-role-of-FTIR-in-protein-analysis-and-biomedical-applications.aspx
- Kaveti, Bhavna. “What are the Advantages and Limitations of Nuclear Magnetic Resonance Spectroscopy?” AZoOptics, 18 Oct. 2023, https://www.azooptics.com/Article.aspx?ArticleID=2460
- Kwan, T. O. C., Reis, R., Siligardi, G., Hussain, R., Cheruvara, H., & Moraes, I. “Selection of Biophysical Methods for Characterisation of Membrane Proteins.” Int J Mol Sci, vol. 20, no. 10, 2019, p. 2605. PMC6566885. doi:10.3390/ijms20102605
- Unraveling the Role of Particle Size Analyzers in Drug Safety, https://www.halolabs.com/blog/unraveling-particle-size-analyzers/
- “Membrane Microscopy is Helping to Ensure That Drugs Are Safe for Patients.” Photonics. https://www.photonics.com/Articles/Membrane_microscopy_is_helping_to_ensure_that/a68087
DISCOVER THE AURA FAMILY
Read More ON this topic