A better way to perform particle analysis
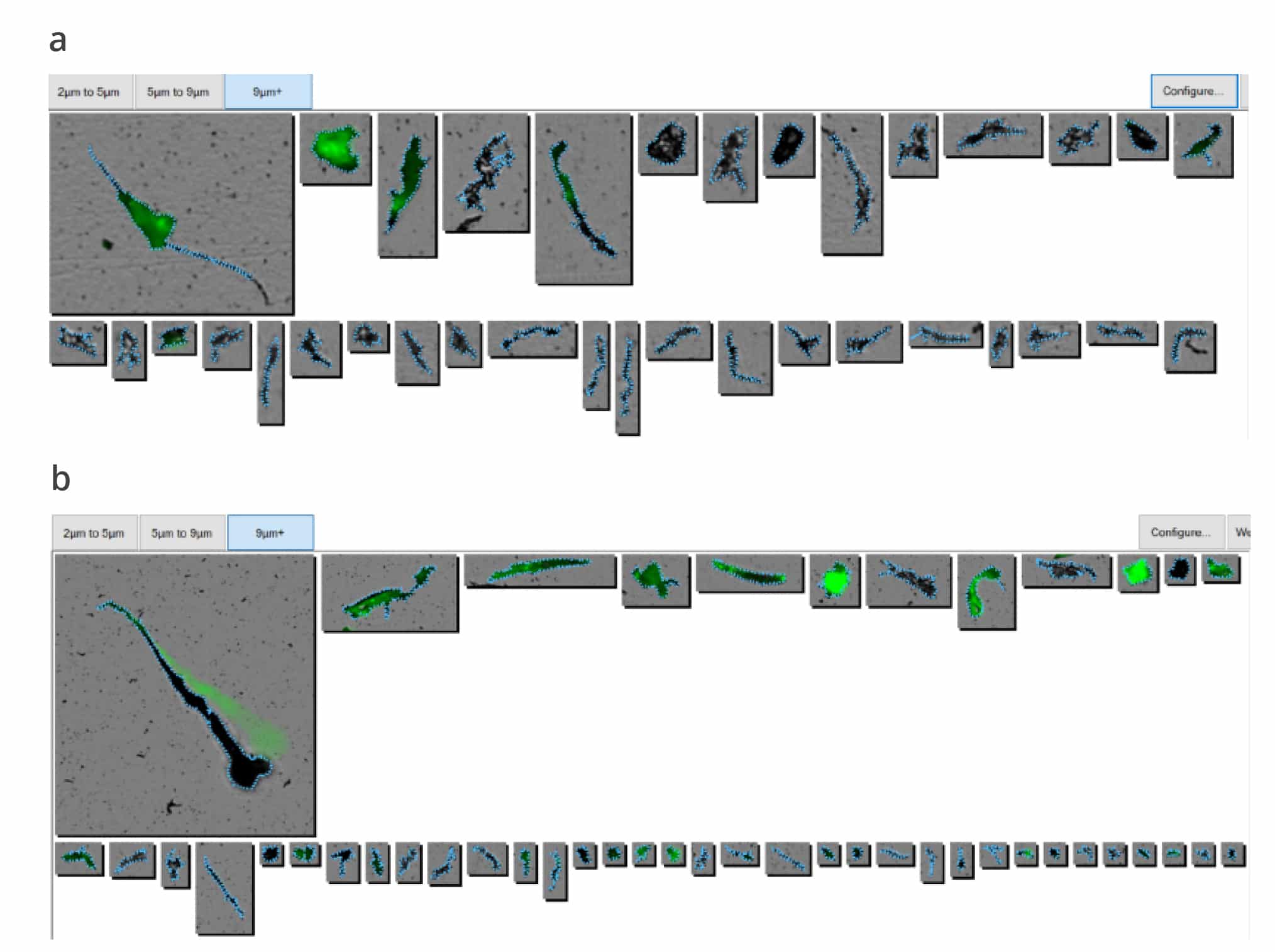
Successful protein therapies require product quality measurements in the late discovery/early development stages that detect, count, and characterize formulation excipients, ensuring protein stability in addition to assessing product stability. But traditional methods require hours of sorting through images, adopting complex machine learning libraries, and often overlook key sample aggregates and degraded compounds. Aura PTx is the first and only system that combines Backgrounded Membrane Imaging (BMI) with two Fluorescence Membrane and Microscopy (FMM) channels to quickly provide count, size, and morphology, and differentiated cellular, protein, or extrinsic aggregates, including formulation excipients.
Why use Aura for Protein Stability Measurement?
Know what is in your sample, faster, easier, and accurately:
- Detect, count, and characterize protein aggregates, formulation excipients, including polysorbate and other degraded products, and subvisible particles
- Get the whole picture with combined Backgrounded Membrane Imaging (BMI) and Fluorescence Membrane Microscopy (FMM) for count, size and identification
- Use just 5 µL of your sample per test, with 100 percent sampling efficiency
- Get results faster with BMI read time of 1 minute/sample, and FMM read time of 90 seconds/sample
- 96-well, high-throughput platform allows comprehensive formulations screening and proper experimental design
- Examine a wide range of particle sizes from 1 µl to 5 mm
- Automate your work with robotics-ready instruments and software
Enhance Your Biophysical Characterizations
By correctly identifying aggregates, degraded excipients, and packaging contaminants, Aura PTx can help you decide if you need to modify formulations or processes and maintain the safety and stability of your protein therapeutics.
All the Data You Need to Determine Protein Stability
Determining your candidate therapeutic’s properties is crucial--aggregation, stability, and hydrophobicity. But traditional methods such as size-exclusion chromatography or dynamic light scattering only capture part of the story.
To fully evaluate proteins for developability assessment and pre-formulations, we need to go beyond traditional techniques and probe into subvisible details that otherwise remain hidden.
Automated analysis with BMI is fast (1 minute/sample) and needs as little as 5 µL per sample. Combined on the Aura PTx with FMM, the techniques can be used early in developability assessment and pre-formulation studies to detect and identify larger, insoluble aggregates.
Featured products
Frequently Asked Questions
Traditionally, high-throughput methods used protein melting points to measure stability, but can only predict stability, not directly measure it. Aura PTx directly measures stability using Thioflavin T (ThT) and BODIPY FL C16 staining to detect and quantify protein aggregates and polysorbate breakdown, respectively. This analyzes sample stability in terms of both biology and polysorbate formulation. Protein stability can be directly measured by quantifying particles formed.
Aura PTx identifies polysorbate and excipient degradation, which impact stability. It also can differentiate protein vs. non-proteins, which also helps develop formulations that increase stability. Aura PTx can also identify native stability of proteins directly without formulations.
Traditional protein stability study methods include SEC and DLS, which fail to capture all aggregates, and microscopy, which is labor-intensive and error-prone. Aura PTx uses BMI to obtain accurate count, size, and morphological information, and FMM to identify subvisible particles and determine protein from non-proteins including aggregates.
The most common parameter for protein stability is Tm, or the point at which half the protein in a sample is unfolded. The higher this figure, the higher the stability. But again, this only predicts protein stability. Moving beyond this traditional technique, Aura PTx Aura accurately and directly measures the effect of formulations on the stability of proteins. Aura PTx determines protein stability by directly characterizing protein state and structure, identifying key aggregates that can contribute to instability.