Get the Stability You Need
The use of lipid nanoparticles (LNPs) as a drug delivery technology is a sophisticated process and requires careful evaluation to ensure their efficacy is not compromised.
As the use of these technologies grows, so does the need for accurate stability measurements.
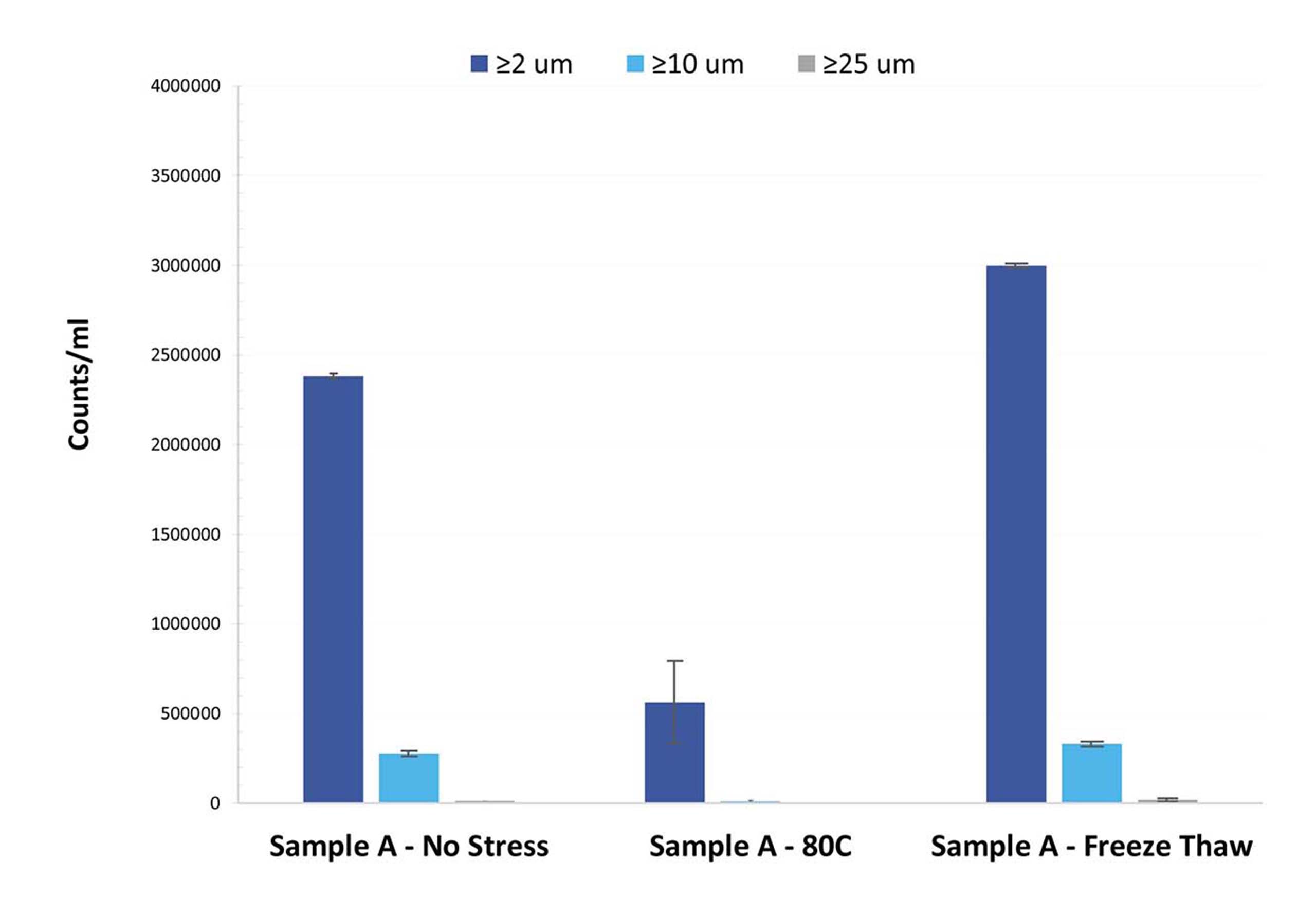
For this reason, low-volume, subvisible particle measurements must be accurately taken to properly assess LNPs and exosomes for stability. However, flow imaging or DLS may not give you the accuracy, speed or flexibility you need.
To help address this critical need, we’re proud to introduce a breakthrough solution: Aura GT, optimized for assessing samples as small as 25 µL for more precise LNP characterization.
Why Use Aura for LNP Characterization and Exosome Characterization?
Because you can achieve so much more insight with much less material:
- Analyze product stability and aggregation analysis
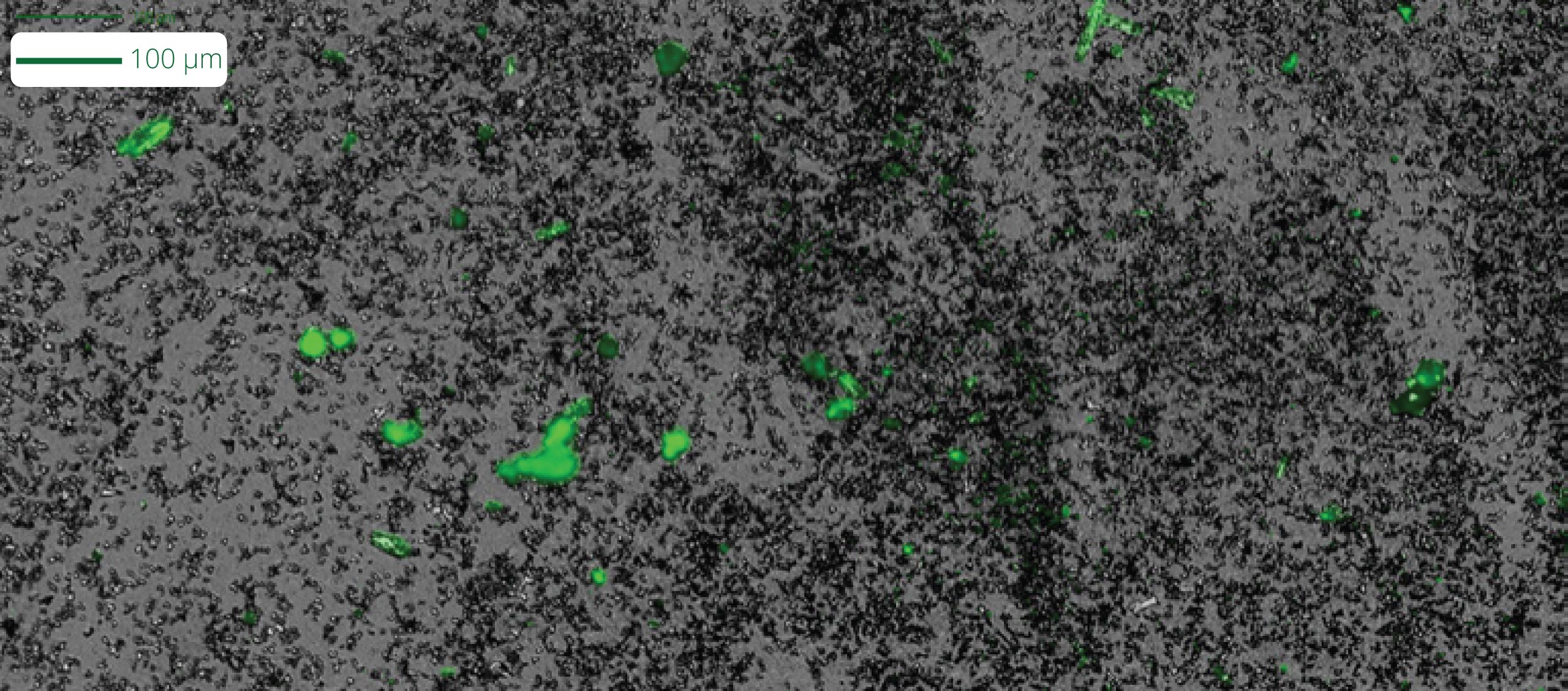
- Identify SYBR™-labeled nucleic acid aggregates from unstable products
- Detect and quantify particles not measured by DLS or SEC
- Determine root cause for unstable gene therapies
- Recommended method by FDA (USP 788 Compendial Method 2)
- Achieve more accurate results with minimal amounts of sample (as little as 5 μL per test)
- Gather comprehensive data and insight into aggregate particles – size, morphology, counts, distribution
- Get insights quickly with rapid analysis time of 1 minute per sample
- Benefit from a wide working range: measure particles from 1 μm to 5 mm with high reproducibility
- Analyze particles without the interference of buffer or matrix for higher sensitivity
- 21 CFR Part 11 software available
Overcome the Challenge of Predicting Quality and Stability of LNPs
Let’s face it: It can be challenging to accurately predict the quality of LNP drugs, even when using samples from well-known commercial developers. Every formulation has its own particle profile, and LNP formulations are no different. This is a common theme in the stability of subvisible particles.
Leveraging our SYBR Gold assay via Fluorescence Membrane Microscopy (FMM), Aura GT solves this issue by assessing both the stability and purity of samples in high throughput, low volume testing, enabling accurate LNP characterization at earlier stages, from initial development all the way through to final product release.
A Smarter and Faster Exosome Characterization Method
Exosome characterization with Aura GT simplifies and expedites the process of selecting the most suitable candidate for development. Much like AAV and LNP therapies, exosomes can aggregate and contain nucleic acids.
Thanks to small sample volumes, stability assessment can occur during earlier development stages, allowing for more informed decisions to be made. This saves valuable time and resources before investing in an unsuitable path that may turn out to be costly and inefficient.
Featured products
Frequently Asked Questions
Lipid nanoparticles can be characterized using various techniques to understand their physical and chemical properties. Common characterization techniques include dynamic light scattering (DLS), which measures the size distribution of nanoparticles in solution; transmission electron microscopy (TEM), providing detailed images of nanoparticle morphology; zeta potential analysis, indicating the surface charge of nanoparticles and their stability in solution; Fourier-transform infrared spectroscopy (FTIR), identifying functional groups within lipid nanoparticles; and differential scanning calorimetry (DSC), analyzing thermal behavior to assess lipid phase transitions and stability. These techniques collectively provide valuable information about the size, shape, surface properties, and stability of lipid nanoparticles, essential for their development and optimization in drug delivery applications.
Solid lipid nanoparticles (SLNs) are nanoscale drug delivery systems composed of lipids that are solid at room and body temperatures. These nanoparticles offer several advantages, including high drug loading capacity, controlled release kinetics, and improved stability compared to traditional drug formulations. SLNs have a spherical morphology with a solid lipid core surrounded by a stabilizing layer, typically composed of surfactants or polymers. They exhibit controlled drug release due to the lipid matrix, which can be tailored to modulate release kinetics. Additionally, SLNs can protect encapsulated drugs from degradation and enhance their bioavailability, making them promising candidates for various pharmaceutical applications such as targeted drug delivery and sustained release formulations. Unfortunately, Aura systems do not currently analyze for SLNs.
Exosomes are small extracellular vesicles released by cells, and their presence and characteristics can be confirmed using specific biomarkers. Commonly used biomarkers for exosome confirmation include tetraspanins (CD9, CD63, CD81), heat shock proteins (HSP70, HSP90), and cell-specific markers (e.g., EpCAM for epithelial-derived exosomes). These biomarkers are typically detected using techniques such as Western blotting, flow cytometry, or immunocytochemistry. By confirming the presence of these biomarkers, researchers can identify and characterize exosomes, which play crucial roles in intercellular communication, biomolecule transfer, and disease pathogenesis, making them valuable targets for diagnostic and therapeutic applications.
Exosome size characterization is essential for understanding exosomes’ biological functions and developing exosome-based therapeutics and diagnostics. Exosomes typically range from 30 to 150 nanometers in diameter, although larger vesicles up to 1000 nm can also be present in some preparations. Size characterization techniques commonly used for exosomes include dynamic light scattering (DLS), nanoparticle tracking analysis (NTA), electron microscopy (EM), and atomic force microscopy (AFM). These techniques provide information about the size distribution, morphology, and concentration of exosomes, facilitating their isolation, purification, and downstream applications in biomedical research and clinical settings. Similar to other biotherapeutics like biologics and proteins, exosomes and extracellular vesicles are also prone to aggregation and instability developing particle sizes ≥1 µm (i.e., subvisible particles).