A Faster Protein Characterization Technique
Developing biologics requires accurate protein characterization, but there hasn't been a reliable way to do this quickly in the past—until now.
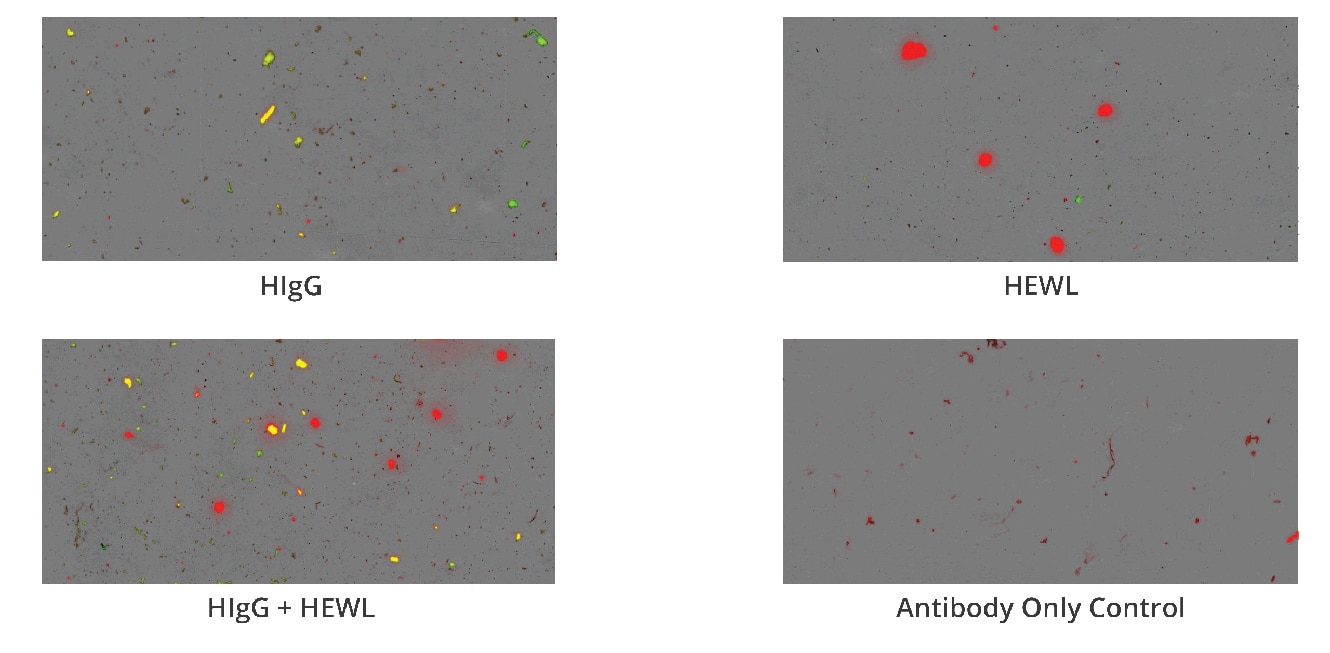
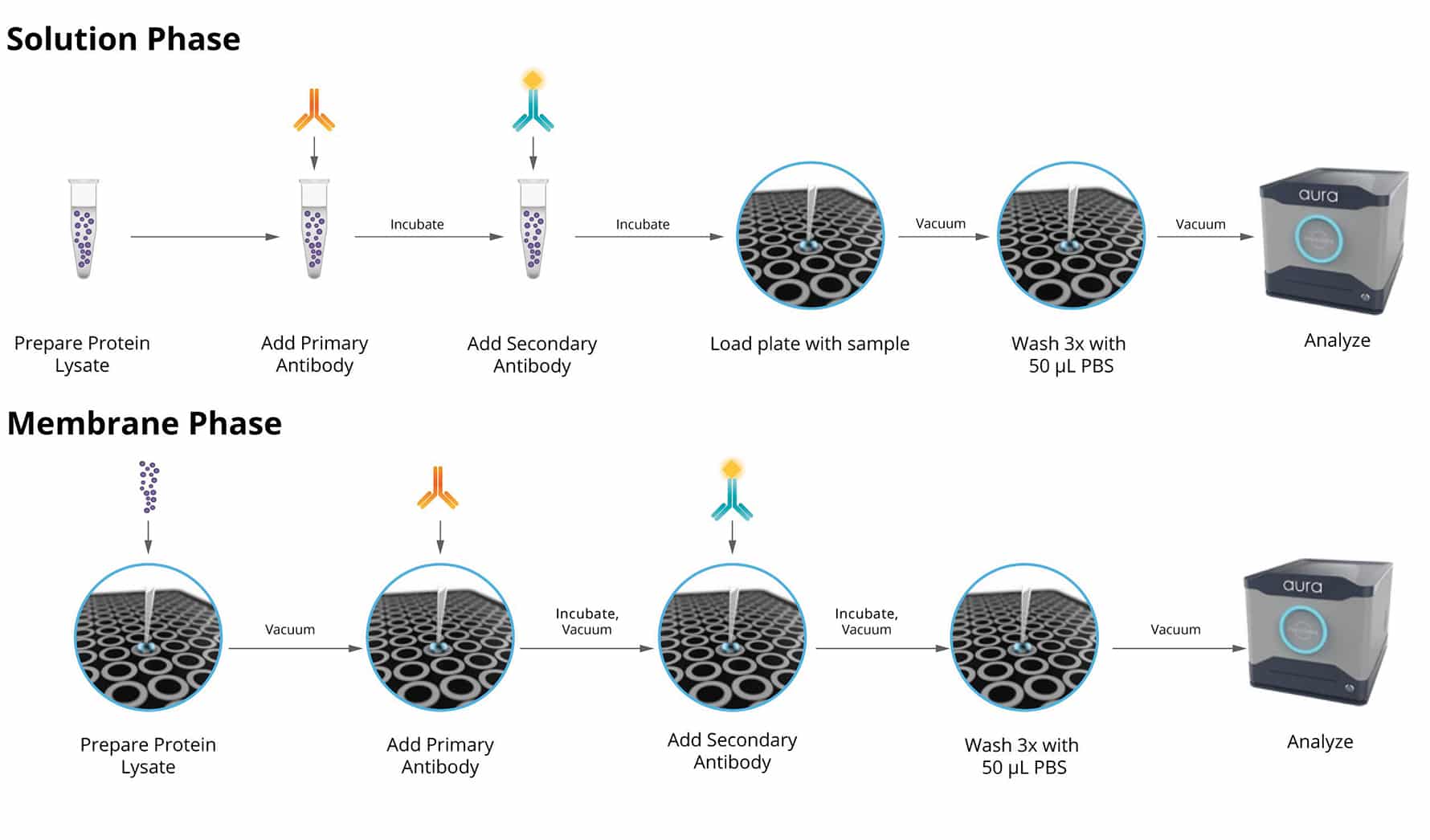
Using the Aura immunoassay, you can quickly pin-point potential instability issues in antibody therapeutics using a two-step fluorescent labeling technique for precise measurement. This multiplexed identification method facilitates faster detection and resolution of any stability concerns. Additionally, it complements other characterization methods to provide a comprehensive analysis of therapeutic stability.
What’s more, Aura immunoassay discerns between two different antibodies (HIgG and BIgG), which is useful for antibody co-formulation applications and assessing drug product versus carrier protein aggregates.
Why Use Aura for Protein Characterization?
Because you can achieve so much more insight with much less material:
- Detect protein aggregates in biologic formulations using fluorescent antibody-based labeling
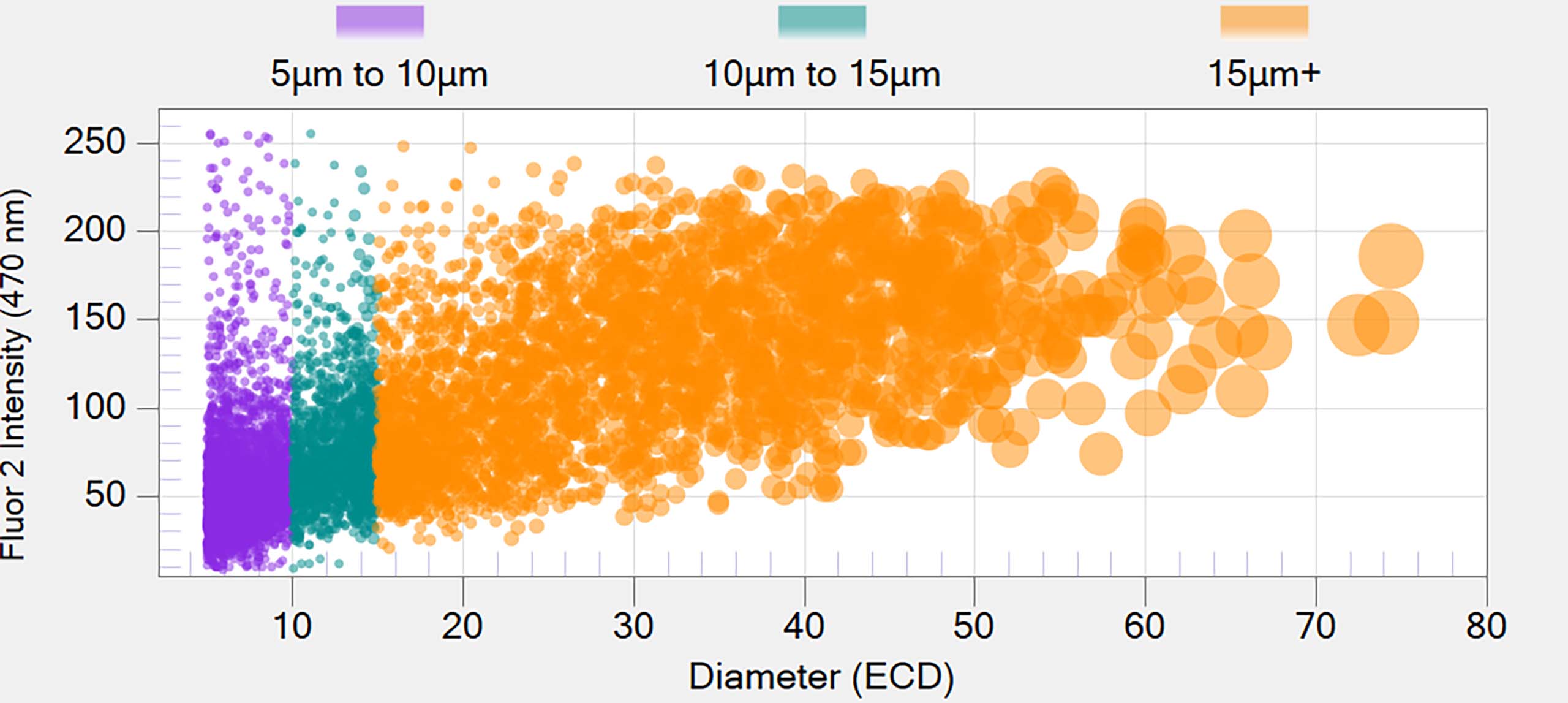
- Obtain protein/non-protein ID for a whole 96-well plate assay down to an individual particle in less than 90 minutes
- Characterize unfiltered cell lines samples through low-volume, high-throughput imaging, counting, sizing and identification
- Identify stable biologic candidates and get high level rankings
- Analyze with only 5 µL to 10 mL of sample
- Quickly analyze data and generate meaningful insights with automated Particle Vue software
- Leverage 21 CFR Part 11 compliant software
Distinguish Proteins from Non-Protein Particles
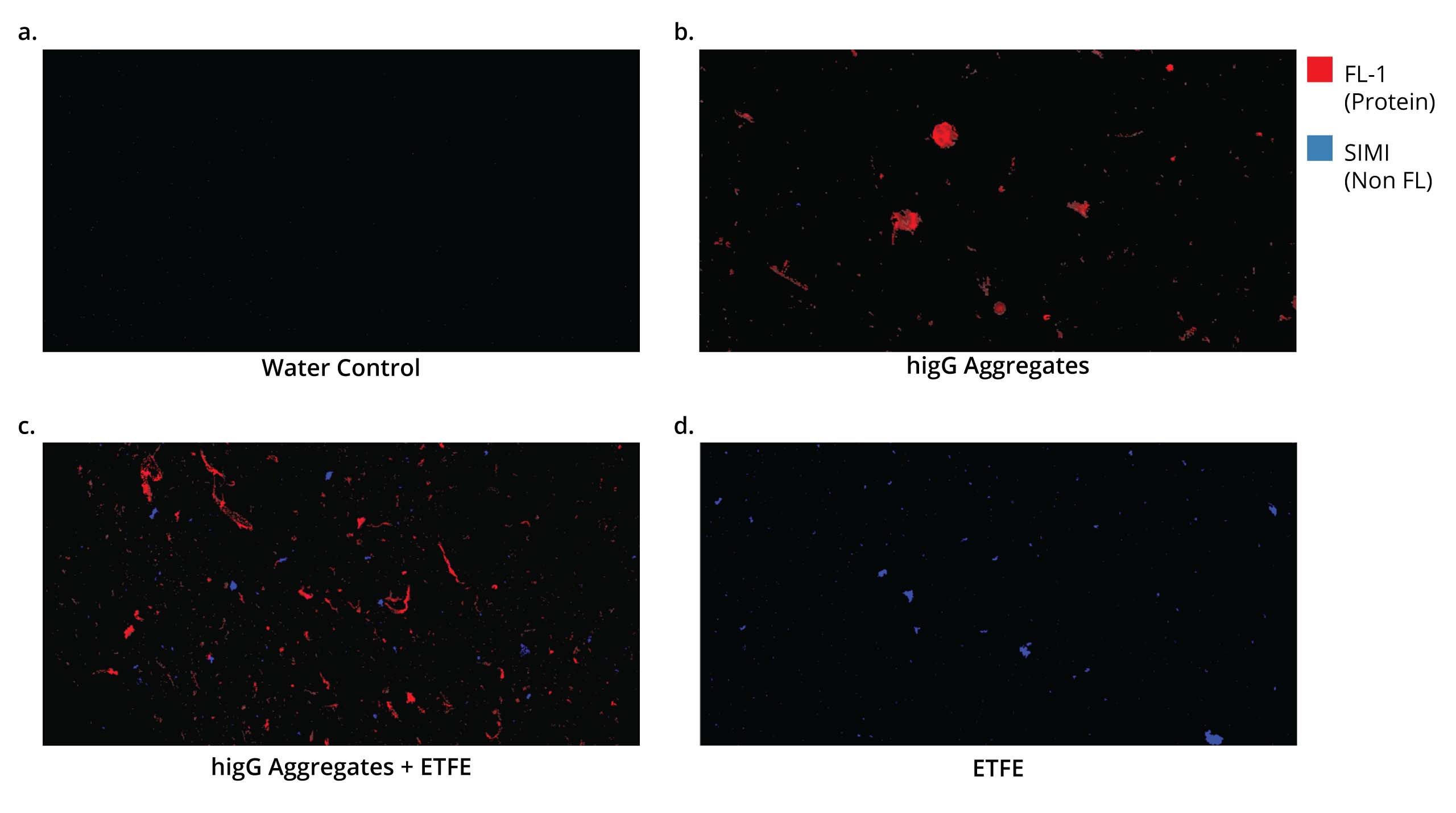
Fluorescence Membrane Microscopy (FMM) , exclusively on Aura PTx, uses Thioflavin T (ThT) dye to conduct high throughput, low volume and specific protein/non-protein particle analysis.
The end result is the ability to obtain protein or non-protein ID for a whole 96-well plate assay down to a single individual particle in less than 90 minutes.
ThT’s high solubility and specificity for protein aggregates makes it possible to differentiate protein aggregates from particles with similar morphology and refractive index, like plastics (e.g., ETFE) and fatty acids.
With 1000x faster throughput compared to spectroscopic methods, FMM brings unprecedented insights into particle characterization.
Accurately Pre-Screen for Antibody Stability
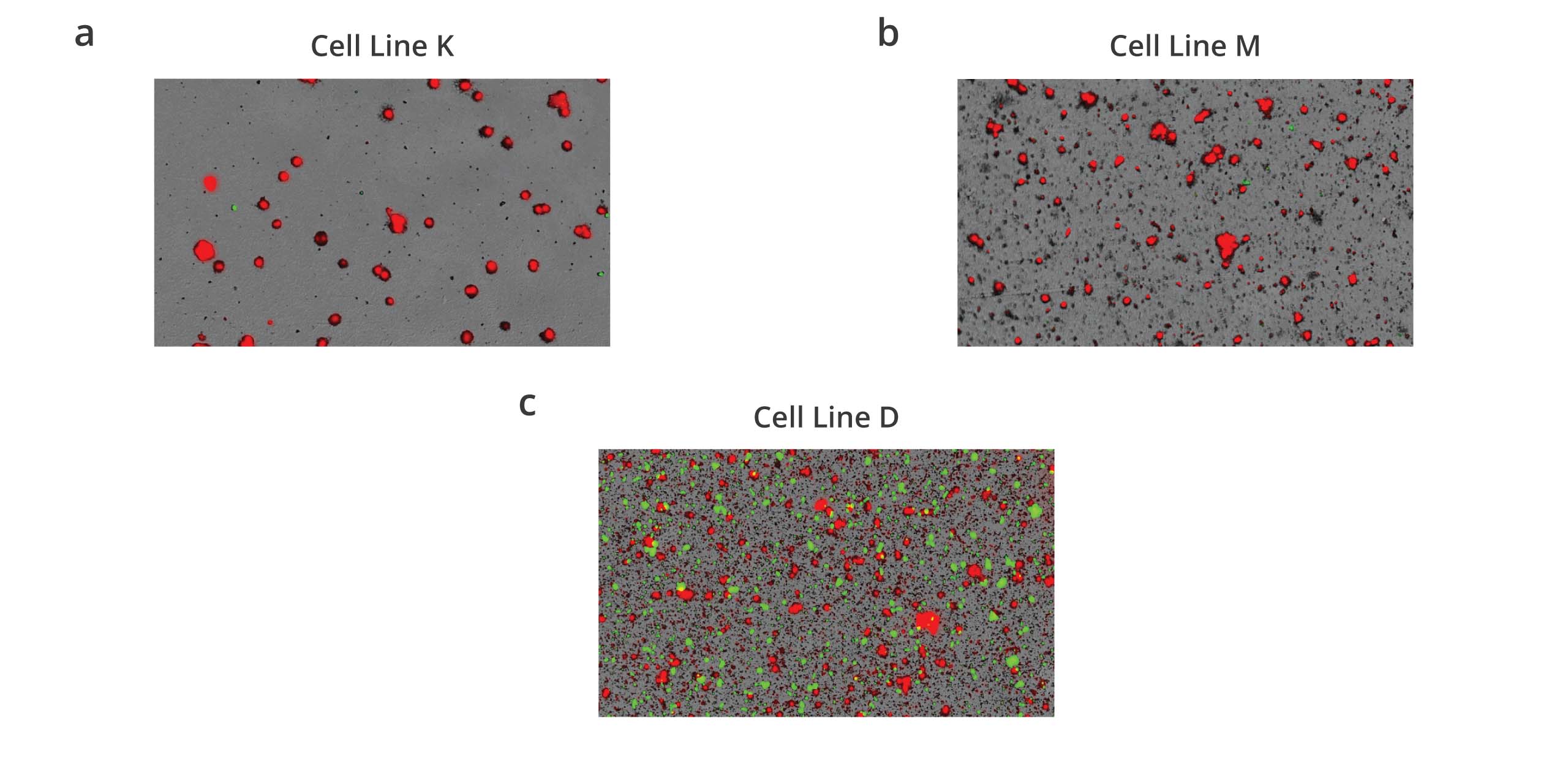
Aura immunoassays bridge the gap between cell line development (CLD) and developability to enable pre-screening for antibody stability once mAbs are secreted from CHO lines.
With Aura immunoassays, stability testing has never been easier. Now you can screen for antibody stability in CHO cells quickly and efficiently using only 40 uL per well. This low-volume method provides positive results within 2 minutes or less, resulting in painless analytical measurements that don’t require entire flasks of cells.
And it’s not just about the immunoassay method - Aura PTx enables quick characterization of unfiltered cell line samples through low-volume, high-throughput imaging, counting, sizing, and identification. Additionally, these techniques can be seamlessly integrated with protein structure characterization to provide a comprehensive analysis of antibody stability.
Analyzing biologically complex cellular and protein samples present in CLD to characterize secreted protein stability has never been faster or more accurate.
Get to the Root of Your Aggregate Source with Particle Vue Software
Aura PTx with Particle Vue software performs high throughput protein characterization and identification of subvisible aggregates across an entire experiment, enabling the ranking of samples based on stability.
The stored images deliver granular insights into the types of physical instabilities found in biologics allowing for deeper understanding and more complete analysis.
Featured Products
Frequently Asked Questions
Protein characterization is the comprehensive analysis of proteins to understand their structure, function, and properties. This process elucidates the primary sequence, higher-order structure, post-translational modifications, interactions, and biological activities of proteins, providing insights into their roles in physiological processes and disease mechanisms. Protein characterization techniques encompass various analytical methods, including chromatography, electrophoresis, mass spectrometry, spectroscopy, and biophysical assays. By characterizing proteins, researchers can assess purity, homogeneity, stability, and immunogenicity, evaluate biosimilarity or comparability, and optimize production and formulation processes for protein-based therapeutics. Protein characterization is essential for ensuring the safety, efficacy, and quality of protein drugs, vaccines, and diagnostics throughout their development and lifecycle.
Proteins require characterization to assess their identity, quality, and suitability for specific applications in research, diagnostics, and therapeutics. Protein characterization provides valuable information about their structure, function, stability, and interactions, enabling researchers and manufacturers to:
- Confirm protein identity: Verifies the primary sequence and structural integrity of proteins, ensuring they match the intended target and meet regulatory requirements.
- Assess purity and impurities: Identifies impurities, contaminants, or degradation products that may affect protein quality, efficacy, or safety.
- Determine structure-function relationships: Elucidates the three-dimensional structure, post-translational modifications, and conformational changes of proteins, correlating with their biological activities and therapeutic effects.
- Evaluate stability and formulation compatibility: Assesses protein stability under various conditions (e.g., temperature, pH, agitation), guiding formulation development and storage conditions to maintain product quality and shelf life.
- Optimize production and purification processes: Optimizes protein expression, purification, and quality control strategies, enhancing yield, purity, and scalability.
- Ensure regulatory compliance: Generates data required for regulatory submissions, demonstrating the safety, efficacy, and quality of protein-based products to regulatory agencies and stakeholders.
Several techniques can be used to characterize proteins, each providing unique information about their structure, function, and properties. Common techniques for protein characterization include:
- Chromatography: High-performance liquid chromatography (HPLC), size-exclusion chromatography (SEC), ion-exchange chromatography (IEC), and affinity chromatography separate proteins based on size, charge, hydrophobicity, or specific interactions, enabling purity analysis and separation of protein isoforms or variants.
- Electrophoresis: Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), isoelectric focusing (IEF), and capillary electrophoresis (CE) separate proteins based on size, charge, or isoelectric point, facilitating purity analysis, subunit composition, and post-translational modifications.
- Mass spectrometry (MS): Liquid chromatography-mass spectrometry (LC-MS) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) identify and quantify proteins, peptides, and post-translational modifications, elucidating primary sequence, structural variations, and interactions.
- Spectroscopy: Ultraviolet-visible (UV-Vis) spectroscopy, fluorescence spectroscopy, circular dichroism (CD), and nuclear magnetic resonance (NMR) spectroscopy allow protein structural analysis as well as analysis of folding, conformational changes, and ligand binding.
- Biophysical assays: Analytical ultracentrifugation (AUC), dynamic light scattering (DLS), surface plasmon resonance (SPR), and differential scanning calorimetry (DSC) characterize protein size, shape, stability, and interactions with ligands or other molecules.
- Immunological assays: Enzyme-linked immunosorbent assay (ELISA), Western blotting, immunoprecipitation, and flow cytometry detect and quantify proteins, epitopes, or specific protein-protein interactions, facilitating immunoassays, biomarker detection, and protein-protein interaction studies.
By integrating multiple techniques, researchers can comprehensively characterize proteins, elucidating their structure-function relationships and guiding their development and applications in various fields.The Aura family of instruments uses a combination of backgrounded membrane imaging (BMI)), a microscopy method and fluorescence membrane microscopy, which uses fluorescent dyes or antibodies for faster, reliable characterization all on one instrument.
For more information on biophysical characterization of proteins, check out our blog post.